Encoding is the process by which a text-based clinical guideline – one that has as its assumed consumer a well-trained clinician – is transformed into a representation that can be read and executed by a computer program.
The SAGE project uses the Protégé knowledge modeling tool, developed at Stanford University, as our encoding workbench. The Guideline Model supplies the building blocks used for encoding by clinical and informatics experts.
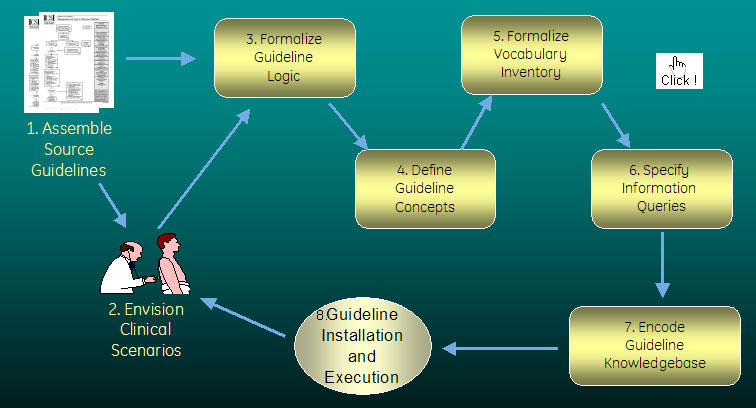
Text-based source guidelines require substantial disambiguation in order to be made computer-interpretable. Specific elements that must be made explicit include the guideline's place in the clinical workflow, the guideline logic, and the medical concepts used. Examples of these elements are detailed in our descriptions of our exemplar guidelines. Guideline encoding is therefore a multi-step process, as diagrammed above and described below.
1. Assemble Source Guidelines |
|

At the start of the guideline encoding process, collaborators collect source materials relating to the guideline. These are ideally textual guidelines and flowcharts produced by expert committees, but may include textbooks, research papers, or other sources. The materials chosen should represent the evidence-based standard of care for the disease process to be managed.
For example, the Diabetes Mellitus guideline encoded by the SAGE project used as its primary source material the American Diabetes Association (ADA) Standards of Medical Care in Diabetes 2006, with additional content derived from the Institute for Clinical Systems Improvement (ICSI) Management of Type II Diabetes Mellitus guideline and The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure.
2. Envision Clinical Scenarios |
|

Evaluation is done to identify opportunities in care workflow for guideline decision support. Appropriate contexts, clinical and administrative events, and desired guideline-based actions must be defined.
Guidelines must be reflected against patterns of care and clinical work plans to identify opportunities for productive intervention. Guidelines should be integrated within an efficient work model, not controlling or distorting work activities and should provide guidance to the individual who is best suited to intervene when intervention is appropriate and timely.
Generally, multi-faceted interventions which reinforce each other across the enterprise are more effective. Physicians are not always the best targets for effective intervention. The recipient community should be educated in the nature and rationale for the guideline; acceptance should be obtained; and the SAGE guideline engine should only support the process identified as ideal. Implementation scenarios will be specific to the organization, the clinical workflow and the capabilities of the information system.
For example, the Community Acquired Pneumonia guideline is executed in the setting of a clinic or ER visit by a patient who is diagnosed with Pneumonia. One important facet of care is timely administration of the first dose of antibiotics. If the physician has already placed the order for antibiotics, and the medication has been requested from the pharmacy but not yet dispensed, the appropriate target for a reminder is the pharmacist. This reminder will be delivered via the most appropriate mechanism enabled by the CIS (e.g. pop-up message, inbox alert, etc.)
For further discussion of the scenarios used for our exemplar guidelines, please see the descriptions of our Immunizations, Diabetes, and Community Acquired Pneumonia guidelines.
3. Formalize Guideline Logic |
|

Each clinical guideline utilizes logic to determine the appropriate course of action (e.g. if a child is 4 months old, and his last Polio vaccine was at least 2 months ago, and he has no allergy to Polio vaccine and no acute illness, he should receive a dose of Polio vaccine). The text-based guideline is distilled by domain experts into a series of logical statements suitable for encoding into a computer-interpretable form.
SAGE generates a written record of the guideline decision logic. This inventory specifies all clinical details required for complete deployment and requires review and integration of all decision elements (contraindications, deferrals, appropriate timing, etc.)
For examples of guideline logic used in our exemplar guidelines, please see the descriptions of our Immunizations, Diabetes, and Community Acquired Pneumonia guidelines.
4. & 5. Define Guideline Concepts and Formalize Vocabulary |
|

Often, concepts present in a text-based guideline require clinical discussion to arrive at an exact definition (e.g. "moderate or severe acute illness" is a precaution for the IPV vaccine). Guideline concepts must be extracted from the guideline logic, contextualized, disambiguated, and de-abstracted. Once clarified and matched into information model requirements, meaning must be reviewed against the appropriate vocabulary domain (e.g. SNOMED CT, LOINC) to assure that the meaning in the guideline corresponds to the meaning to be retrieved from the patient record.
For examples of specific concept inventories for our exemplar guidelines, please see the descriptions of our Immunizations, Diabetes, and Community Acquired Pneumonia guidelines.
For a detailed discussion of concept representation, please see Controlled Resources.
6. Specify Information Queries |
|

The SAGE Guideline Engine must obtain patient data from the CIS to perform logic, and every CIS represents patient data differently. Interoperable decision support must be able to interact correctly with any vendor information model. Since we cannot tell CIS vendors how to structure their systems, the SAGE approach to interoperability is to use a standard information model – the Virtual Medical Record, or VMR – with the understanding that each vendor will build their own translation from the standard to their system.
7. Encode Knowledgebase |
|

The encoding process culminates with the use of a "guideline workbench" to encode an electronic version of the guideline. The SAGE project uses Protégé, an open-source knowledge modeling tool developed at Stanford University. Tools available with Protégé provide encoding assistance, such as highlighting logical inconsistencies or workflow 'dead ends'. It also provides access to standard vocabularies for the population of concept structures in the encoded guideline.
The ideal guideline encoder has both clinical experience and technical expertise, as encoding is a fairly advanced exercise in both of these areas. Collaboration between technical and clinical experts can be a good approach.
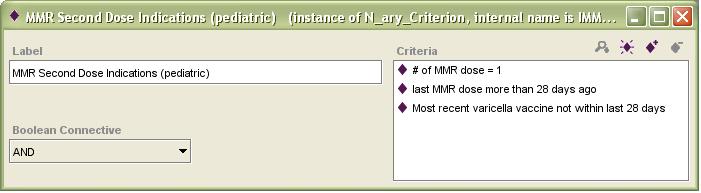
Even a simple-seeming clinical guideline should be expected to take weeks or months to encode, depending on the specific guideline and the experience level of the encoder(s). For example, the Immunizations guideline is, superficially, a straightforward schedule for recommended vaccine administration. However, a vast array of relative and absolute contraindications and reasons for deferral must be represented and encoded. Making up missed immunizations is also a complex matter, as many make-up recommendations vary based on an interplay between the timing of previous immunizations, the need for other immunizations, and the age of the patient now and at the time of prior immunizations. For example, here is a screen capture of the Protégé-based representation of the indications for the second pediatric dose of MMR vaccine:
Several software components are needed before the encoding process may begin. If you are interested in installing a SAGE encoding environment, please see our downloads page.
8. Install and Execute Guideline |
|

An encoded guideline must be verified and tested for errors prior to deployment. The SAGE Tab provides a Protégé-based tool for this purpose. When testing in Protégé, you can enter patient data by hand or from files, or you may turn-on a live connection to a clinical information system (CIS) to obtain patient data.
Following Protégé-based testing, a guideline may be installed and tested in the context of a functioning CIS (CIS). Prior to execution, the encoding must be tailored to the specific capabilities and data representation strategies of the CIS. Often, the guideline must also be altered to reflect the practice conventions of the local site at which the CIS is used.
An important step in the localization of a guideline is the binding of standard concept representations to the specific forms used by the local CIS. See Deployment. The SAGE tool set includes tools for mapping. This mapping process must be carried out by an individual with knowledge of both standard and local representations. The table below shows an example mapping from a standard LOINC code for a lab test to the four representations of that test used at one of our test installation sites, The Mayo Clinic. Since the second and third concepts are used only in research and will never appear in a patient's record, the mapping need not be done for those two local concepts.
| LOINC Code | Local Code Name | Local Code | Performed / Reported |
|---|---|---|---|
| 4548-4 | Hemoglobin A1C,B | 82080 | Central Clinical Lab/In CIS |
| 4548-4 | Hemoglobin A1C,B | 80947 | Clinical Trials/Not in CIS |
| 4548-4 | Hemoglobin A1C,B | 124055 | New England Research/Not in CIS |
| 4548-4 | Hemoglobin A1C,B | 82990 | Peripheral Clinical Lab/In CIS |
To date, SAGE guidelines, due to their status as a NIST research project, have never been deployed in an active patient care system.
For further details on guideline installation and execution, please see the Deployment section of this web site.

 top
top